Example datasets
Table of contents
Exemplar data for testing MCMICRO
Note that the representative images above depict only a subset of the image channels present in each data set.
Two exemplars are currently available for demonstration purposes:
exemplar-001is a minimal reproducible example for running all modules of the pipeline, except the dearray step.- The exemplar consists of a small lung adenocarcinoma specimen taken from a larger tissue microarray (TMA), imaged using CyCIF with three cycles.
- Each cycle consists of six four-channel image tiles, for a total of 12 channels.
- Because this exemplar is too small for automated illumination correction, illumination profiles were precomputed from the entire TMA and included with the raw images.
exemplar-002is a two-by-two segment of a TMA that is meant to test the dearray step, followed by processing of all four cores in parallel. .- The four cores are two meningioma tumors, one GI stroma tumor, and one normal colon specimen.
- This data set has ten cycles with 40 total channels and also includes precomputed illumination profiles.
After installing Nextflow, both exemplars can be downloaded using the following commands:
nextflow run labsyspharm/mcmicro/exemplar.nf --name exemplar-001 --path /local/path/
nextflow run labsyspharm/mcmicro/exemplar.nf --name exemplar-002 --path /local/path/
(Content download to the directory indicated by
/local/path/.)
Or, the examples can be downloaded as .zip files.
Note: When downloading the example files from this link, your system will need enough space for both the .zip file and for the expanded contents (indicated above). Downloading via these links is slower than the command line download, but may be preferred by users that are not using Nextflow.
- Exemplar-001: https://mcmicro.s3.amazonaws.com/exemplars/exemplar-001.zip - 240 MB (320 MB unzipped)
- Exemplar-002: https://mcmicro.s3.amazonaws.com/exemplars/exemplar-002.zip - 2.5 GB (3.5 GB unzipped)
Visual Guide: Processing Exemplar-002 with MCMICRO
This detailed visual guide walks you through the MCMICRO pipeline steps as it processes exemplar-002. This guide was generated using the Minerva software package.
Want to give it a try?? Process Exemplar-001 and -002 using our tutorial!
Exemplar Microscopy Images of Tissues (EMIT)
The EMIT (Exemplar Microscopy Images of Tissues) data set is reference data containing a variety of images that can be used to develop and benchmark computational methods for image processing. These data are expected to be useful for biological studies; however, annotation of this data set for biological interpretation ongoing. Users of these data should expect to see updates and additions - these will be versioned and made backwards compatible whenever possible.
Presently, EMIT comprises one tissue microarray (TMA) data set and one whole-slide image (WSI) data set. Users of these resources should cite Schapiro et al 2021.
Tissue microarrays (TMAs)
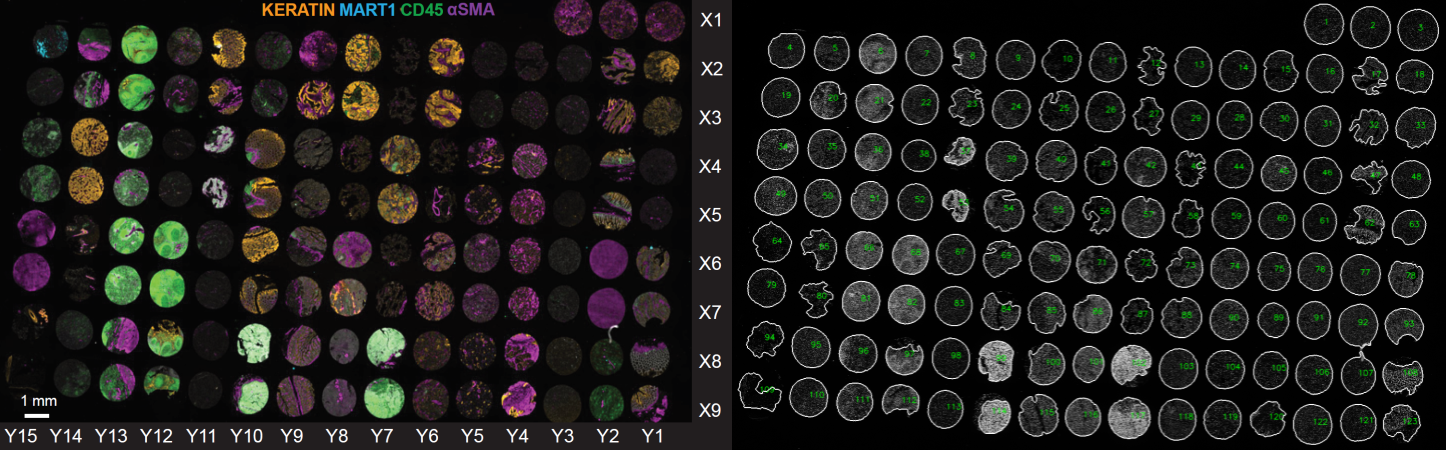
The TMA contains cores from 34 cancer, non-neoplastic diseases, and normal tissues collected from clinical discards under an IRB-supervised protocol. The TMA was imaged using the cyclic immunofluorescence (CyCIF) method described in Lin et al 2018. Data were collected with a 20X magnification, 0.75 NA objective with 2x2-pixel binning using two multiplex antibody panels (on section 11 “TMA11” and section 22 “TMA22”).
Whole-slide images (WSIs)
Data was collected from a tonsil specimen (4 year-old female, caucasian). The specimen was serially sectioned with each section processed using a different imaging techonology. The resulting images comprise a 100+GB dataset, which has been processed by MCMICRO with primary data and all intermediates available on Synapse.