
Great....yet another TMA dearray program. What does this one do?
Coreograph (code) uses UNet (Ronneberger et al., 2015), a deep learning model, to identify complete/incomplete tissue cores on a tissue microarray, and export them individually for faster downstream processing. It has been trained on 9 TMA slides of different sizes and tissue types.
Training sets were acquired at 0.65 microns/pixel resolution and downsampled 1/32 times, or 25 times, to speed up performance. Once the center of each core has been identifed, active contours is used to generate a tissue mask of each core that can aid downstream single cell segmentation. A GPU is not required but will reduce computation time.
Troubleshooting scenarios
NOTE for HMS users: When using Coreograph on O2, the O2tma profile should be used!!
1. Alright, let’s get started!
When using MCMICRO, Coreograph does not require any additional input parameters to run. The DNA channel is assumed to be in the 1st channel.
To enable Coreograph, add tma: true to the workflow namespace of your params.yml:
workflow:
tma: true
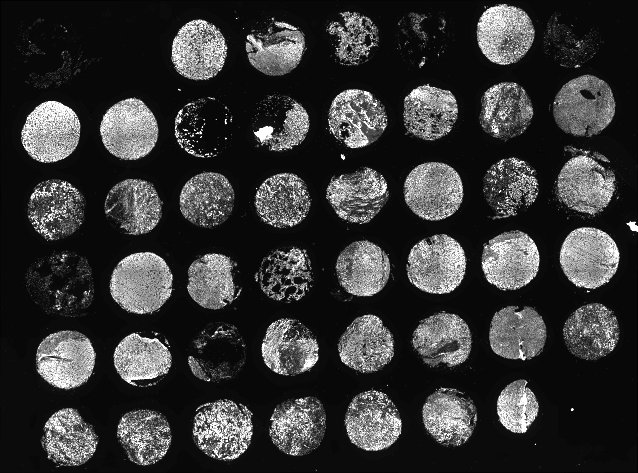
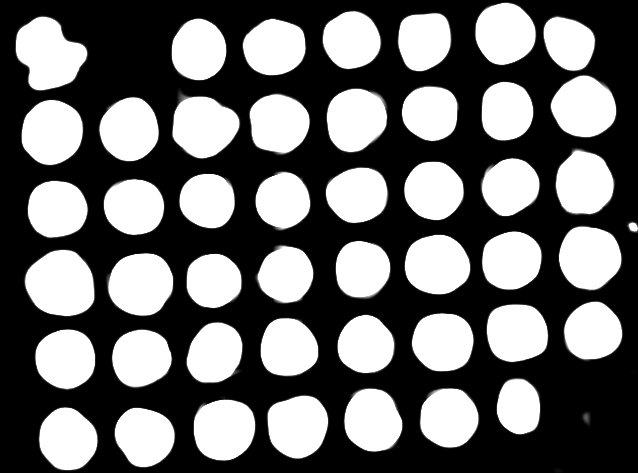
As one can see, each core is labelled with a single number implying that each core was found uniquely. Furthermore, each core has a thick white line to indicate the accuracy of segmenting each core. (Future versions will have a colored outlines for better visibility).
2. Well, my DNA channel is not in the 1st channel.
No problem! Specify --channel in the coreograph field of module options. The field uses 0-based indexing, i.e., the 1st channel has index 0. For example, to use the 4th channel, your params.yml may look as follows:
workflow:
tma: true
options:
coreograph: --channel 3
3. The cores aren’t being found properly.
Coreograph is trained on various core sizes ranging from 500 microns to 2 mm acquired at a pixel size of 0.65 microns per pixel and then downsampled 5 times. If your core size or image resolution are significantly different, you will need to either upsample or downsample a different number of times using --downSampleFactor. See below for examples:
3a) If your pixel size is 0.325 microns per pixel, then your pixel size is double the training data by a factor of 2 (0.65/0.325). You should downsample more times. Use 6 instead of 5.

Corresponding params.yml:
workflow:
tma: true
options:
coreograph: --downsampleFactor 6
3b) If your pixel size is 1.3 microns per pixel, then your pixel size is half of the training data (0.65/1.3). Instead of downsampling by 5 times (default), you should downsample less. Try 4.

Corresponding params.yml:
workflow:
tma: true
options:
coreograph: --downsampleFactor 4
4. What do the square outlines indicate?

The white square indicate that a core was found but Coreograph could not detect the exact outlines. A possible cause is dim staining. Downsizing 4 times instead of 5(default) can be a potential solution.
Corresponding params.yml:
workflow:
tma: true
options:
coreograph: --downsampleFactor 4
Note: The line segments inside the circle outlines are an artefact of how Coreograph draws the outlines. They do not indicate any issues.

