"So... you want to do single-cell image segmentation?"
Troubleshooting scenarios
1. I’m new to this whole segmentation thingy. And I have a deadline. Just get me started with finding nuclei!
In its simplest form, s3segmenter by default will identify primary objects only (usually nuclei) and assumes this is in channel 1 (the first channel). In this case, no settings need to be specified.
If you specified segmentation-channel in your workflow parameters, the channel index will be correctly propagated to s3seg and any preprocessing steps. For example, if your params.yml was as follows:
workflow:
segmentation: unmicst # Can be omitted, since this is default
segmentation-channel: 5
then channel 5 will be passed to both UnMICST and S3Segmenter to ensure that the two steps remain in sync.
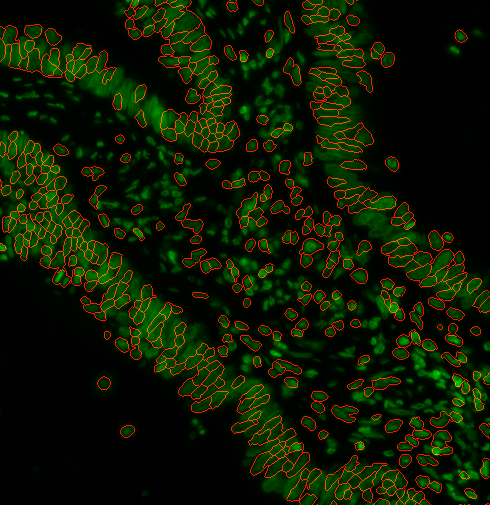
2. It’s a disaster. It’s not finding all the nuclei
Depending on the type of pre-processing that was done, you may need to use a different method of finding cells. Let’s add --nucleiRegion localMax to the options:
options:
s3seg: --nucleiRegion localMax
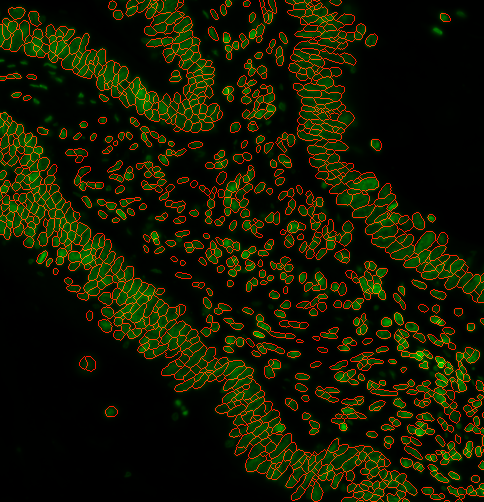
3. Looks good! I want to filter out some objects based on size
You can specify a range of nuclei diameters that you expect your nuclei to be. Using --logSigma <low end of range> <high end of range> Ie. --logSigma 10 50 will retain all nuclei that have diameters between 10 and 50 pixels. Default is 3 60
Examples: a)
options:
s3seg: --nucleiRegion localMax --logSigma 3 10
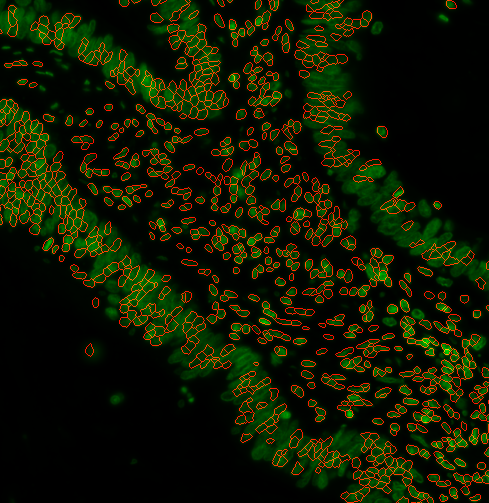
b)
options:
s3seg: --nucleiRegion localMax --logSigma 30 60
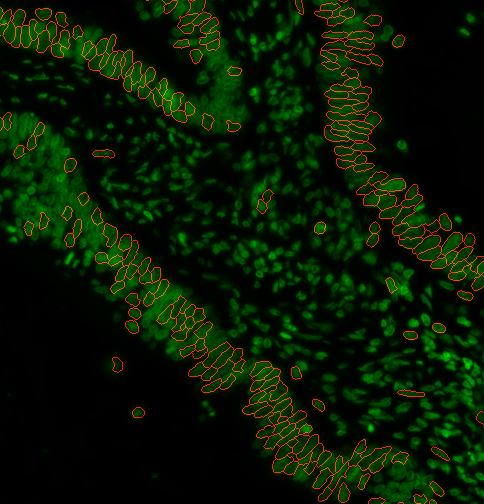
c)
options:
s3seg: --nucleiRegion localMax --logSigma 3 60
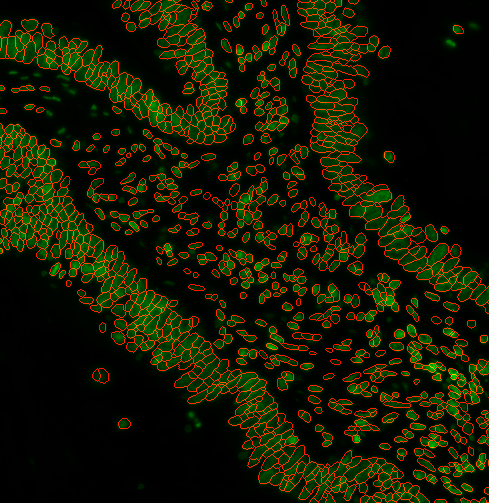
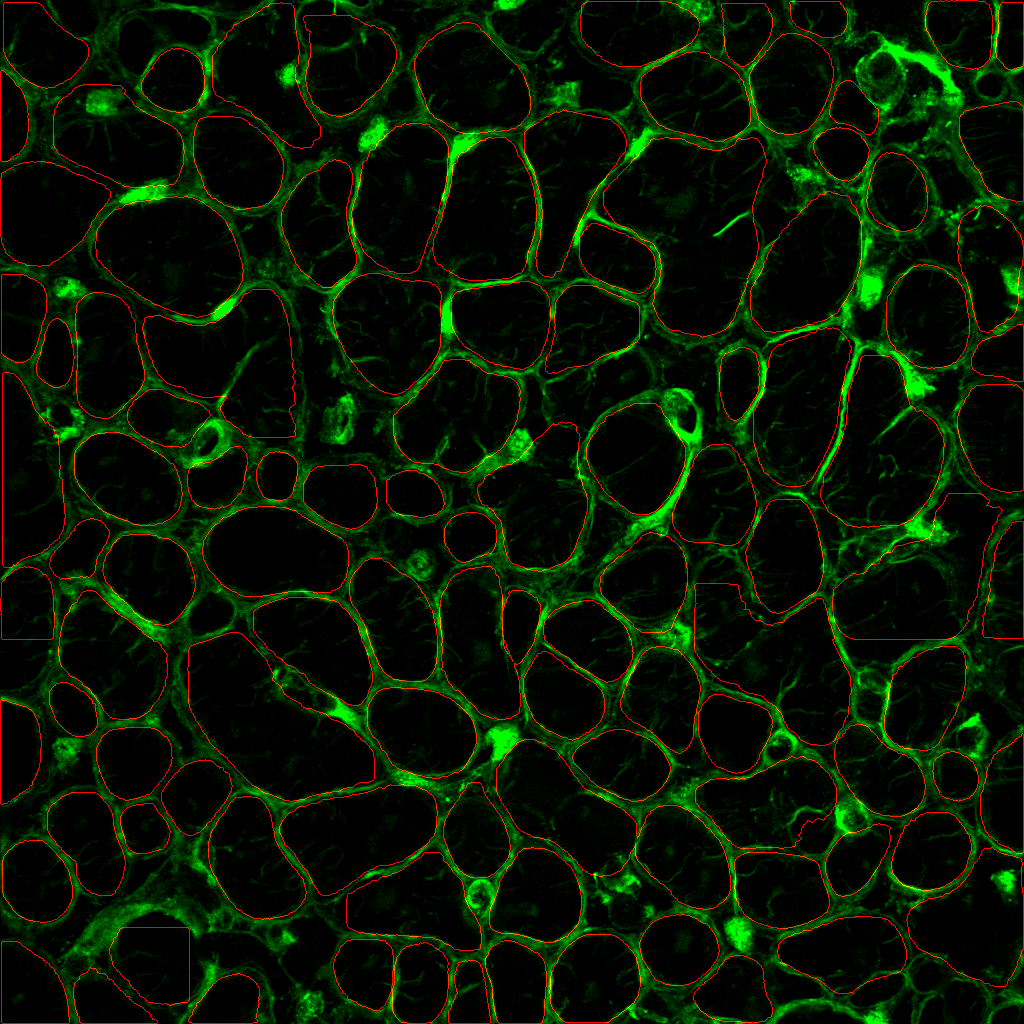
4. a) How do I segment the cytoplasm as well?
To do this, you will need to:
- look at your image and identify a suitable cytoplasm channel such as the example below.
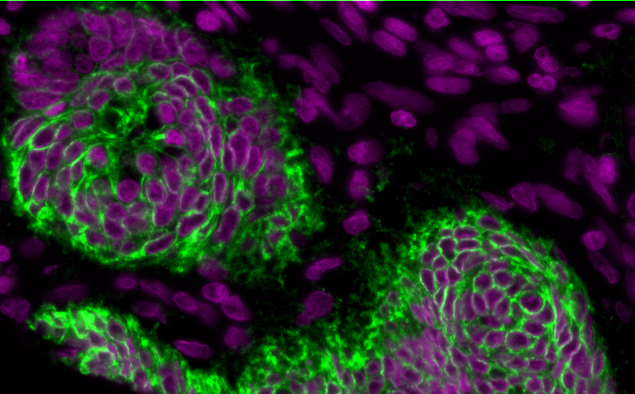
Nuclei and cytoplasm stained with Hoechst (purple) and NaK ATPase (green) respectively. Notice how the plasma membrane is distinctive and separates one cell from another. It also has good signal-to-background (contrast) and is in-focus.
Specify --CytoMaskChan <channel number(s) of cytoplasm>. For example, to specify the 10th channel, use --CytoMaskChan 10. To combine and sum the 10th and 11th channels, use --CytoMaskChan 10 11. Doing this maximizes the ability to capture more cells.
- Also, specify this to activate cytoplasm segmentation:
--segmentyCytoplasm segmentCytoplasm
Putting it together in a params.yml file may look as follows:
options:
s3seg: --nucleiRegion localMax --CytoMaskChan 10 --segmentCytoplasm segmentCytoplasm
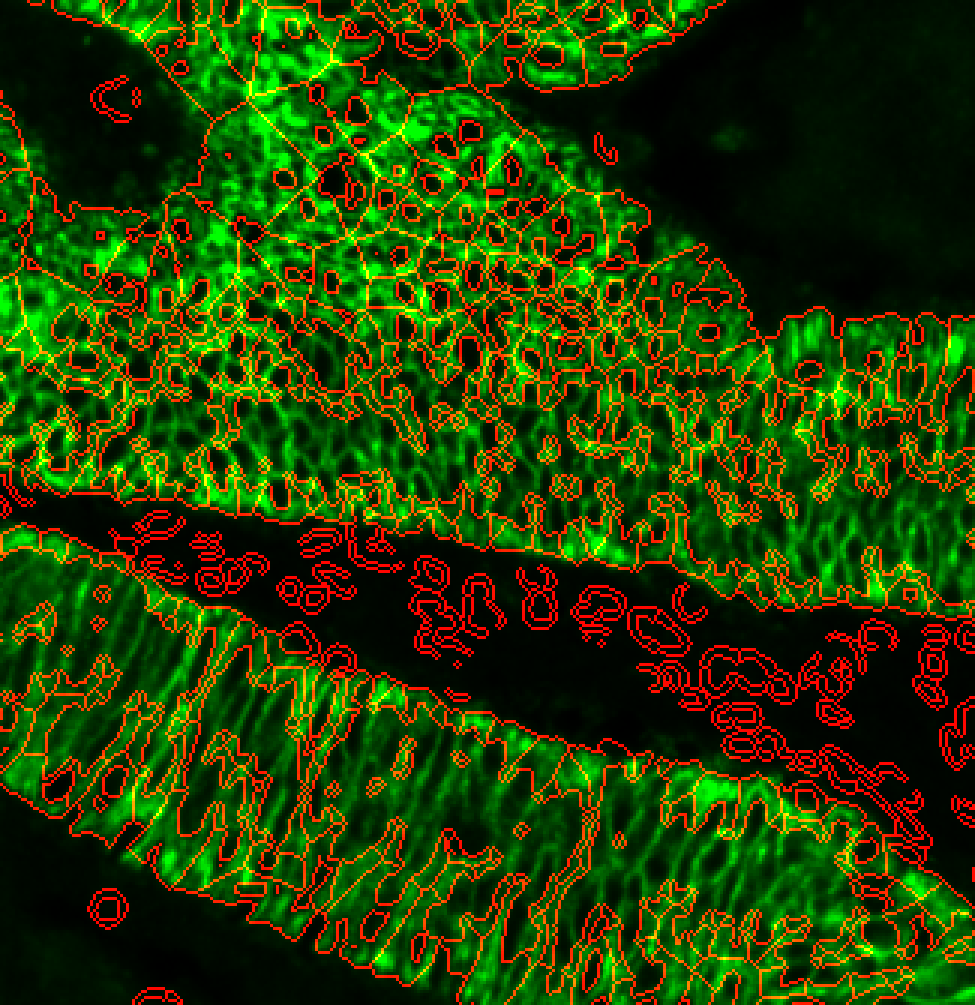
4. b) I don’t have a suitable cytoplasm channel…..
That’s ok. Cytoplasm segmentation is hard because there isn’t a universal marker. It’s generally acceptable to sample some number of pixels around the nucleus to approximate the cytoplasm.
- Choose
--cytoMethod ring - Then, specify the width of this ring
--cytoDilation <thickness of ring in pixels>ie.--cytoDilation 3will surround the nuclei with a 3-pixel thick cytoplasmic ring. The default is 5 pixels.
Examples
i)
options:
s3seg: --nucleiRegion localMax --CytoMaskChan 10 --segmentCytoplasm segmentCytoplasm --cytoMethod ring --cytoDilation 15
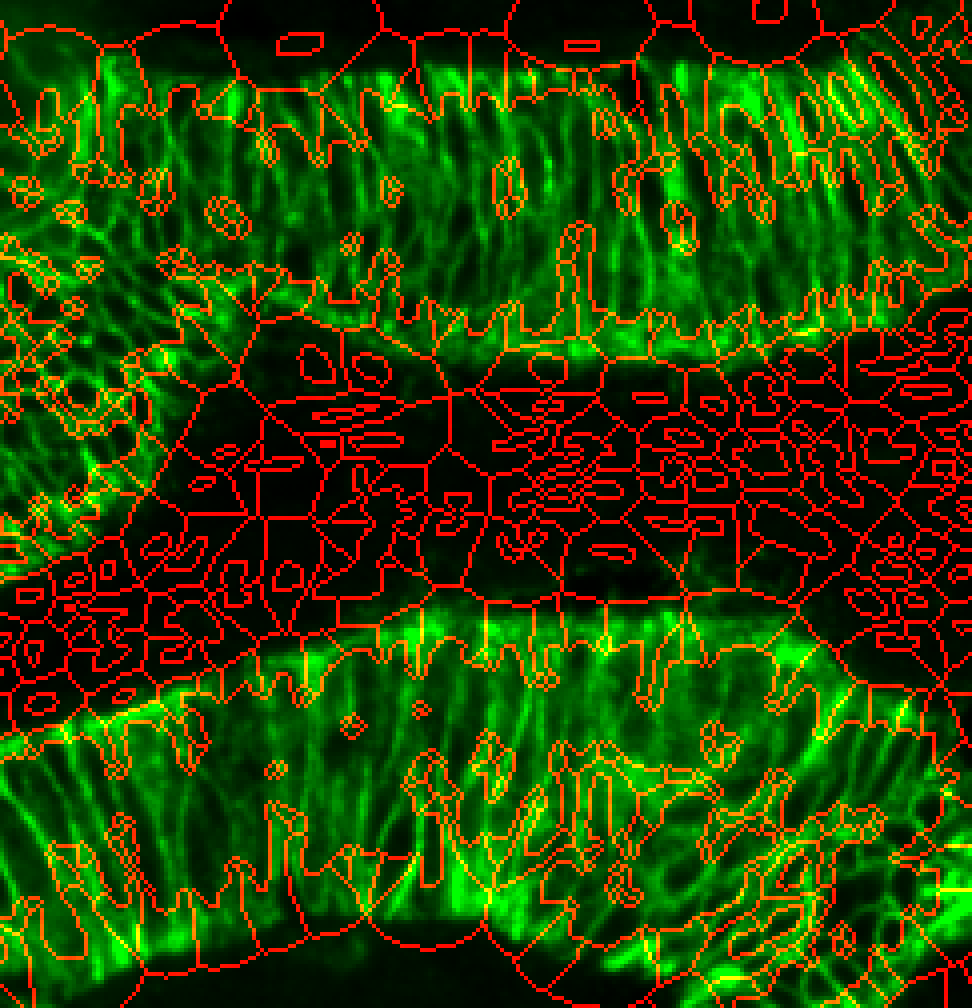
Cytoplasm spilling beyond cytoplasm stain. Possibly too large --cytoDilation parameter.
ii)
options:
s3seg: --nucleiRegion localMax --CytoMaskChan 10 --segmentCytoplasm segmentCytoplasm --cytoMethod ring --cytoDilation 6
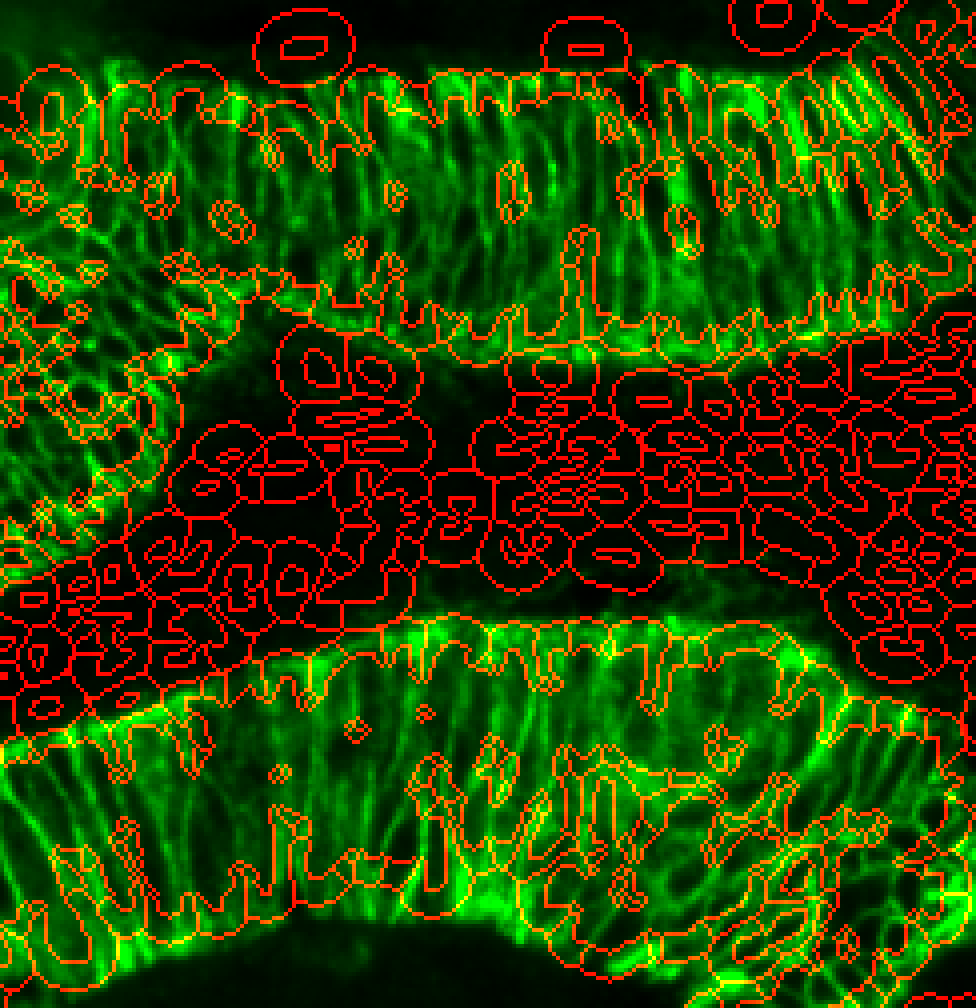
Much better. Cytoplasm outlines now just within the marker stain.
4. c) Are there other ways to detect the cytopolasm?
There’s a hybrid approach that combines a cytoplasm channel and the ring around the nuclei to deal with tissues that have sporadic cytoplasm staining. Try changing –cytoMethod to ‘hybrid’.
options:
s3seg: --nucleiRegion localMax --CytoMaskChan 10 --segmentCytoplasm segmentCytoplasm --cytoMethod hybrid
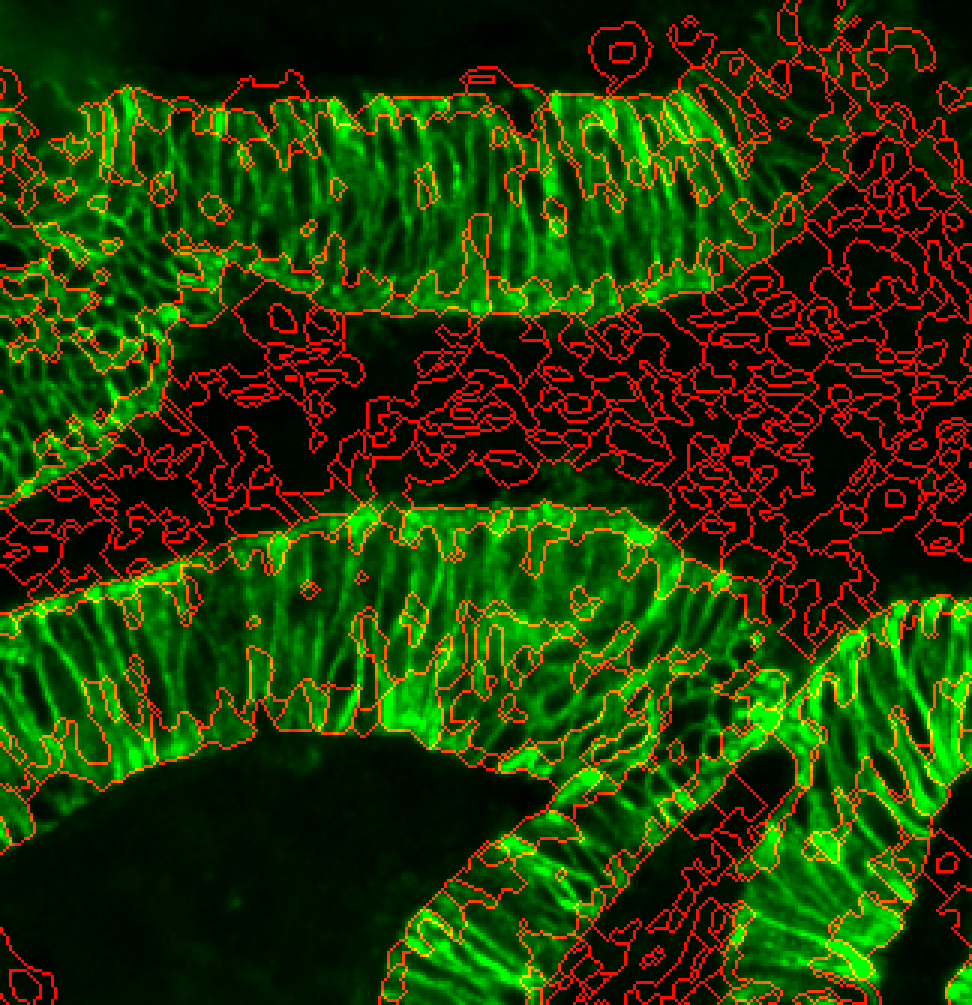
This is still a very experimental technique and may not yield better results!
5. I have an instance segmentation model, which already produces a mask. How do I incorporate this in?.
S3segmenter can accept pre-made instance segmentation primary object masks and still run some of the later functions we talked about above. To bypass nuclei segmentation, specify --nucleiRegion bypass. Then, you can still use --logSigma to filter overly small/large objects.
options:
s3seg: --logSigma 45 300 --nucleiRegion bypass
6. Nuclei…. Cytoplasm… NOW GIVE ME INTRACELLULAR SPOTS
This is a very complexed operation and requires several parameters.
Detail which channels you want to run spot detection on.
--detectPuncta <channel number(s)>. ie.--detectPuncta 1 2 3will look for spots in the 1st, 2nd, and 3rd channels.--punctaSigma <the sigma(s) of the spots in each channel>. This is equivalent to the standard deviation of a fitted Gaussian curve through one of your spots. Select custom values for each channels. Ie.--punctaSigma 1.5 2 1.75If you specify one value, it will use that for all channelsLastly, choose the sensitivity of the spot detector.
--punctaSD <standard deviations for each channel>.
Lower numbers increase sensitivity at the expense of more false positives. Not advisable when images are very noisy. May need to work out SD empirically.
Select custom values for each channels. i) ie. --punctaSD 10 12 10 (more stringent) Note: If you specify one value, it will use that for all channels
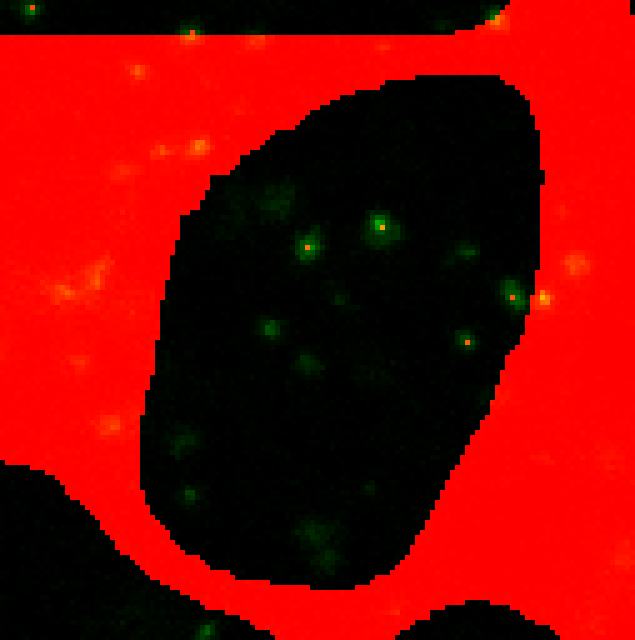
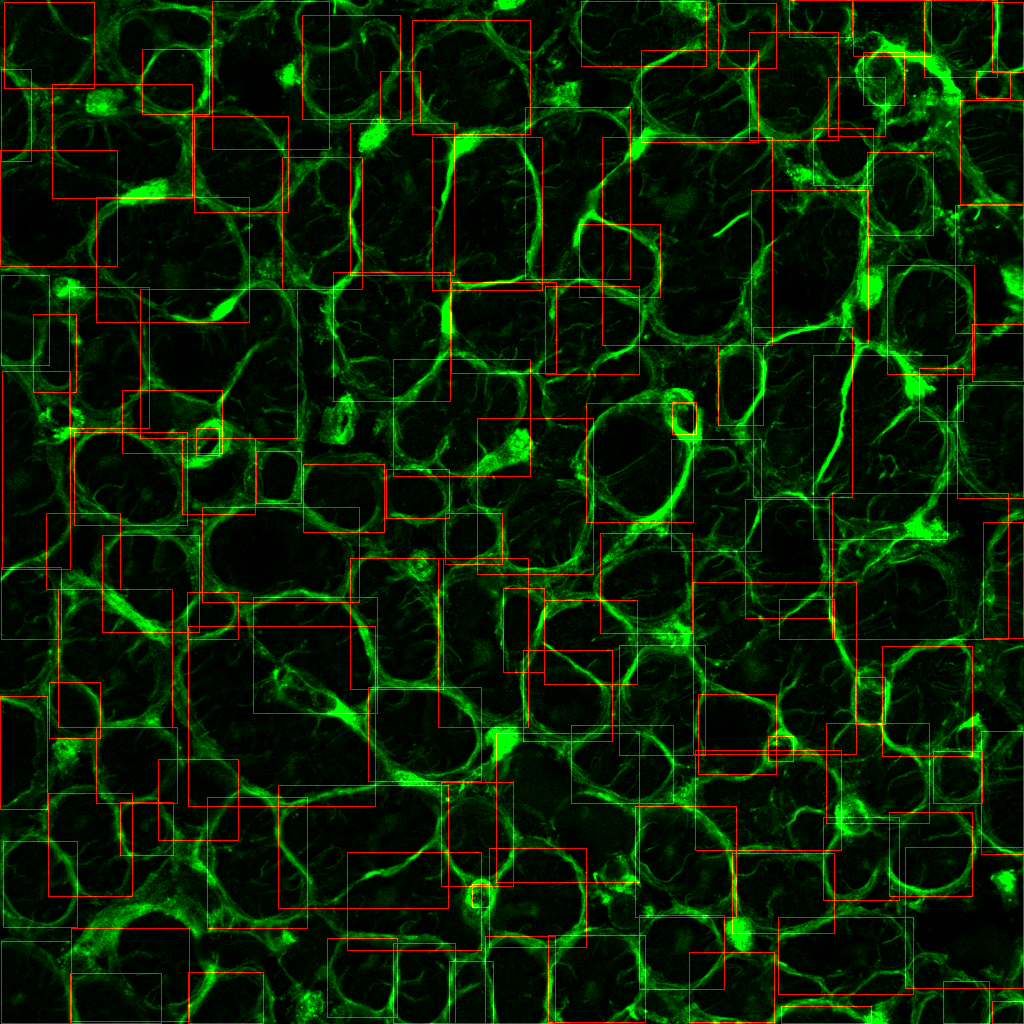
Hmmm….only 4 puncta are being detected shown by the green dots, but there are clearly other spots not being picked up.
ii) --punctaSD 3 3 3 (more sensitive)
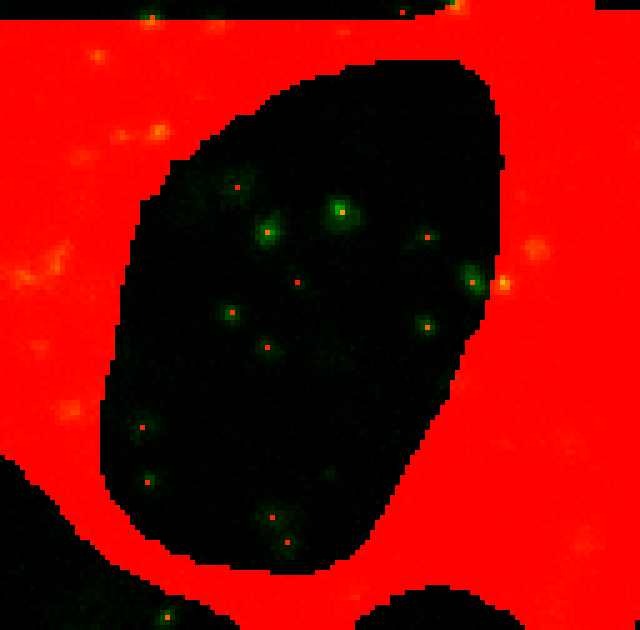
Perfect! All visible spots appear to be detected with the more sensitive option.
options:
s3seg: --logSigma 45 300 --detectPuncta 1 2 3 --punctaSigma 1.5 2 1.75 --punctaSD 3

