UnMICST - Universal Models for Identifying Cells and Segmenting Tissue

(pronounced un-mixed)
Introduction
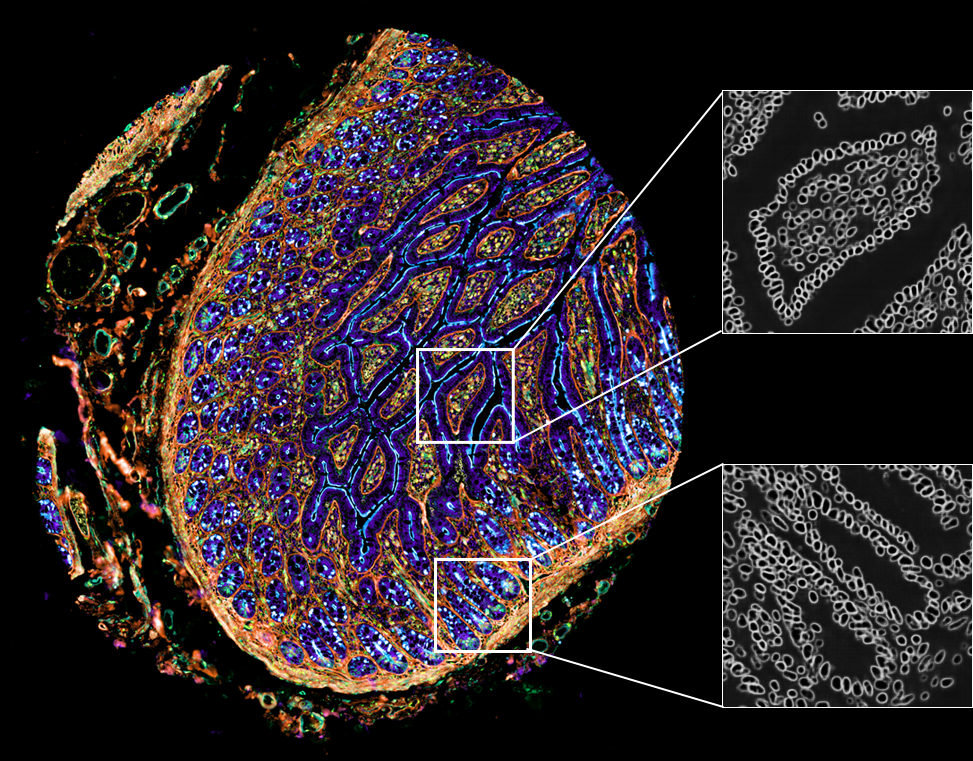
Nuclei segmentation, especially for tissues, is a challenging and unsolved problem. Convolutional neural networks are particularly well-suited for this task: separating the foreground class (nuclei pixels) from the background class. UnMICST generates probability maps where the intensity at each pixel defines how confident the pixel has been correctly classified to the aforementioned classes. UnMICST can utilize a nuclear envelope channel (lamin B and nucleoporin 98) alongside a DNA channel to improve segmentation accuracy, in addition to more standard single-channel segmentation.
These probability maps can make downstream image binarization more accurate using tools such as S3segmenter (Saka et al., 2019). UnMICST currently uses the UNet architecture (Ronneberger et al., 2015) but Mask R-CNN and Pyramid Scene Parsing (PSP) Net are coming very soon!
Training data
Quality machine learning algorithms can only be generated from quality training data.
UnMICST has been trained on 6 issue types that encapsulate the different morphologies of the entire tissue microarray: 1) lung adenocarcinoma, 2) non-neoplastic prostate 3) non-neoplastic small intestine, 4) tonsil, 5) glioblastoma, and 6) colon adenocarcinoma.
UnMICST also trained on real augmentations - such as intentionally de-focused planes and saturated pixels - to further improve segmentation accuracy relative to real-world experimental artifacts.
For more information about accessing the training data visit: https://github.com/HMS-IDAC/UnMicst
or read the publication here: (Yapp et al., 2021)
Troubleshooting Scenarios
1. I just wanted to get started.
Set --tool unmicst-solo in the unmicst field of module options, and choose a channel that has your DNA stain in the segmentation-channel field of workflow parameters. Channel specification uses 1-based indexing. An example params.yml may look as follows:
workflow:
segmentation-channel: 1
options:
unmicst: --tool unmicst-solo

2. My tissue images have very packed nuclei. What do I do??
unmicst-solo uses a single DNA channel whereas unmicst-duo uses a DNA channel and a nuclear envelope stain, which can help the model discriminate between tightly-packed nuclei. This additional stain can come from markers such as lamin B1, B2, nucleoporin 98 or some additive combination. Set --tool unmicst-duo and choose channels that have your DNA and nuclear envelope stains. If your DNA and envelope stains are in the 1st and 5th channel respectively, the corresponding params.yml may look as follows:
workflow:
segmentation-channel: 1 5
options:
unmicst: --tool unmicst-duo

3. My tissue images are blurry. What do I do??
Again, consider using unmicst-duo with a nuclear envelope stain.
without nuclear envelope stain

with nuclear envelope stain

4. You said the training data is sampled at 0.65microns/pixel and acquired with a 20x/0.75NA objective lens. What do I do if my data was acquired with a 40x lens?
First of all, check what is your pixel size since that is more relevant. If your pixel size is about half of the training data (ie. 0.325 microns/pixel), use a --scalingFactor of 0.5. If your pixel size is double (ie. 1.3 microns/pixel), then set your --scalingFactor to 2. For example,
options:
unmicst: --scalingFactor 2
5. I heard unmicst-legacy is spectacular.
You mean spectacularly bad. unmicst-legacy is deprecated. Use unmicst-solo or unmicst-duo if you have a nuclear envelope staining.

